Restoring a linear function with a single neuron¶
Artificial neuron¶
In the previous article we talked about a model that has a \theta, set of parameters, which we need to optimize in order to minimize the J(\theta) loss function. When it comes to deep learning, such models are actually artificial neural networks, consisting of nodes - neurons.
Usually an artificial neuron is mathematically represented as some nonlinear function from a single argument (a linear combination of all input signals) the signal of which is sent to a single output. Figure 1 shows a diagram of an artificial neuron:
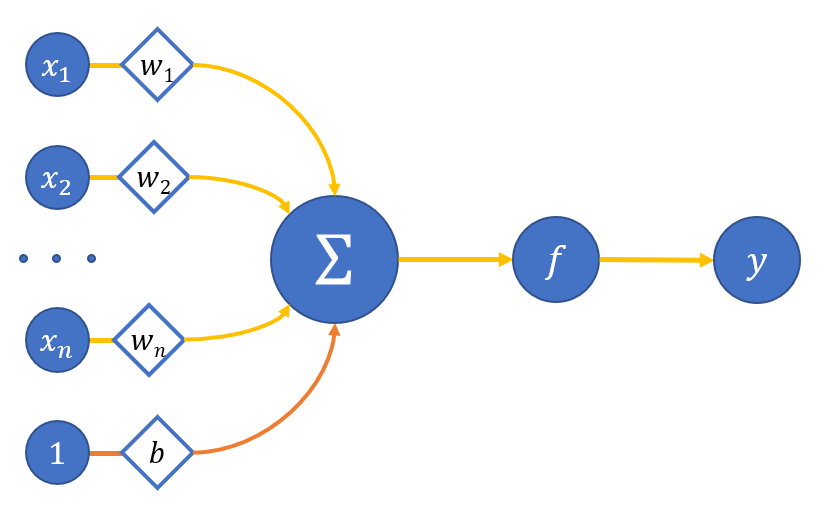
where
x_0,...,x_n - neuron inputs;
w_o,..w_n - weights of the corresponding inputs;
b - bias weight (we equal the input of this connection to one);
\sum - adder of weighted inputs;
f - neuron activation function;
y - neuron output.
As a result, the output of the neuron looks like this:
or, if you set the bias weight as w_0 and the input value is x_0:
A different activation function could provide an output of 1, if the linear combination of all neurons exceeds a certain value, or 0 if vice versa.
If we take such neuron for the model previously discussed, then its weights will be the exact parameters \theta of the model.
We will take a model of a simplified neuron with one input and no activation function (yet) for convenience - figure 2:
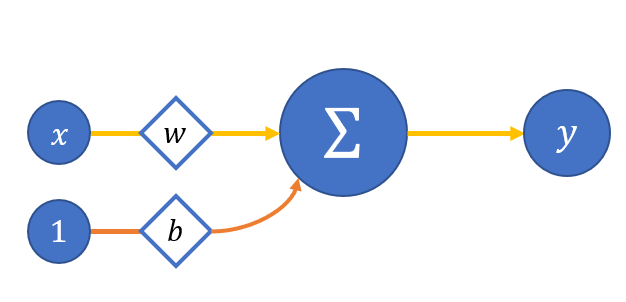
Then:
which reminds the equation of a straight line a lot, so let us restore it.
To do this we have to solve the optimization problem:
We take the mean squared as a loss function (two is added in the denominator for convenience of the further differentiation):
where
N - the number of objects in the dataset;
y_i - the real value for the i-th object;
y_i^p - the value predicted by the model for the i-th object.
A little reminder on how we optimize the parameters:
or in case of a simplified neuron:
How do we find \nabla_{w}{J_i(w_t, b_t)} and \nabla_{w}{J_i(w_t, b_t)}? All you need is to take the partial derivatives of the loss function from these parameters, which will also be the derivatives of a complex function:
We have the following analytical expressions for the selected loss function:
Then update the artificial neuron parameters:
Time to implement all this in code.
A simplified neuron in code¶
First, we import all that we need - this is numpy for working with tensors,matplotlib.pyplot for plotting graphs and write the functions show andshowSubplots for displaying graphs on the screen:
import numpy as np
import matplotlib.pyplot as plt
def show(x, y, pred=None, title=None):
plt.plot(x, y, linewidth=5, color="black", antialiased=True, label="True values")
if pred is not None:
plt.plot(x, pred, linewidth=2, color="orange", antialiased=True, label="Predicted values")
if title is not None:
plt.title(f'{title}')
plt.legend()
plt.show()
def showSubplots(x, y, *args, title=None):
fig = plt.figure(1, figsize=[10.4, 4.8])
for i, arg in enumerate(args):
i += 1
ax = fig.add_subplot(int("12{}".format(i)))
ax.plot(x, y, linewidth=5, color="black", antialiased=True, label="True values")
yp = arg["y"]
name = arg["name"]
color = arg["color"]
ax.plot(x, yp, linewidth=2, color=color, antialiased=True, label="{}".format(name))
ax.legend()
if title is not None:
fig.suptitle(f'{title}')
fig.show()
x = np.linspace(-3, 3, 1000, dtype=np.float32).reshape(-1, 1)
def func(x):
return 2 * x + 3
f = np.vectorize(func)
Y = f(X)
show(X, Y)
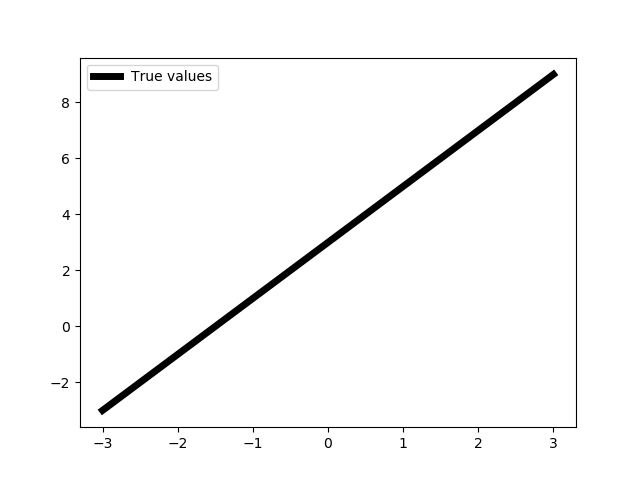
We do not really need np.float32 data type for now, but we will need it later.
Now we will create a simple implementation of the selected loss function - the mean squared error:
class Error:
@staticmethod
def value(true, pred):
return 0.5 * np.mean((true - pred) ** 2)
@staticmethod
def grad(true, pred):
c = 1 / np.prod(true.shape)
return -(true - pred) * c
c - a coefficient of the average inverse to the number of objects in the dataset.
Finally, the artificial neuron class:
class Neuron:
def __init__(self):
self.w = 1
self.b = 0
self.inData = None
self.data = None
self.grad = None
def __call__(self, data):
return self.forward(data)
def forward(self, data):
self.inData = data
self.data = data * self.w + self.b
return self.data
def backward(self, grad):
self.grad = grad
def update(self, lr=0.1):
self.w -= self.inData * self.grad * lr
self.b -= self.grad * lr
def optimize(self, data, target, lr):
prediction = self(data)
print("Neuron error {}".format(Error.value(target, prediction)))
grad = Error.grad(target, prediction)
self.backward(grad)
self.update(lr)
A bit of information on the methods implemented:
forward- a forward propagation of data through the neuron; we need to store the input data in the class attributes here, because it will be needed during the parameters optimization (see x in the weight update formula above);backward- something that we will need in the future, for now we just save the gradient that comes from the loss function in the class attributes;update- updating the neuron parameters using the aforementioned formula;optimize- a method that combines all the necessary operations to optimize the parameters of a neuron;lr- learning rate.
Now we can move on to the neuron training.
The neuron training¶
We need to initialize the neuron and compare the values it outputs with the desired function:
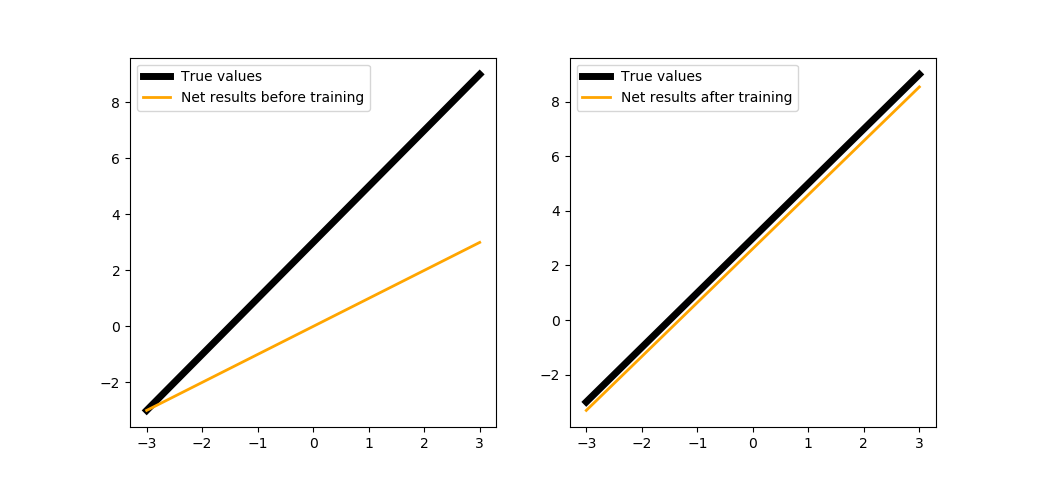
def trainNeuron(steps=200, learnRate=1e-2):
neuron = Neuron()
predictedBT = [neuron(x) for x in X]
for i in range(steps):
idx = np.random.randint(0, 1000)
x = X[idx]
y = f(x).astype(np.float32)
neuron.optimize(x, y, learnRate)
predictedAT = [neuron(x) for x in X]
showSubplots(
X,
Y,
{
"y": predictedBT,
"name": "Net results before training",
"color": "orange"
},
{
"y": predictedAT,
"name": "Net results after training",
"color": "orange"
}
)
When training in 200 steps:
trainNeuron(200)
We could train the neuron a bit longer:
trainNeuron(500)
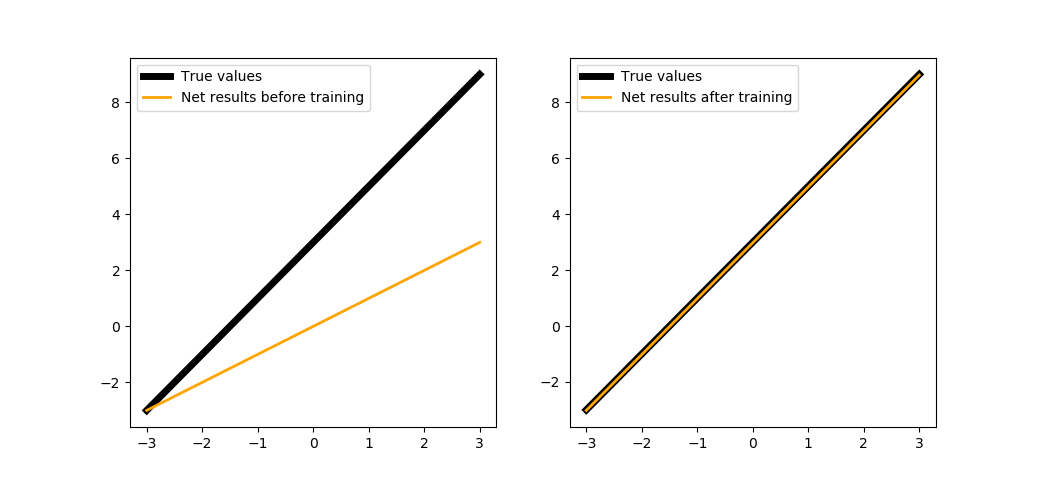
The next section will show how this was implemented in PuzzleLib.
Implementing the library tools¶
Now we will create a separate function to parallel the training of our neuron with the training of neuron built by the library's tools.
We will need a linear layer module through which we will simulate a neuron, an optimizer, a loss function, and a function for placing tensors on the selected device (usually that would be GPU) to_gpu to work with training a neuron on PuzzleLib:
def trainBoth(steps=1000, learnRate=1e-2):
from PuzzleLib.Modules import Linear
from PuzzleLib.Optimizers import SGD
from PuzzleLib.Cost import MSE
from PuzzleLib.Backend.gpuarray import to_gpu
Next, we declare the function that will optimize our pseudo-neuron:
def optimizeModule(module, cost, optimizer, data, target):
module.trainMode()
data = to_gpu(data.reshape(-1, 1))
target = to_gpu(target.reshape(-1, 1))
error, grad = cost(module(data), target)
print("PL module error {}".format(error))
module.zeroGradParams()
module.backward(grad, updGrad=False)
optimizer.update()
A data reshape is necessary because PuzzleLib imposes certain requirements on the dimensions of the input data.
Let us create a pseudo-neuron and fill in the values of weights and biases so that they coincide with the neuron we created:
neuronPL = Linear(insize=1, outsize=1)
neuronPL.W.fill(1)
neuronPL.b.fill(0)
Next, we initialize the loss function and the optimizer, setting the latter to a pseudo-neuron:
cost = MSE()
optimizer = SGD(learnRate)
optimizer.setupOn(neuronPL)
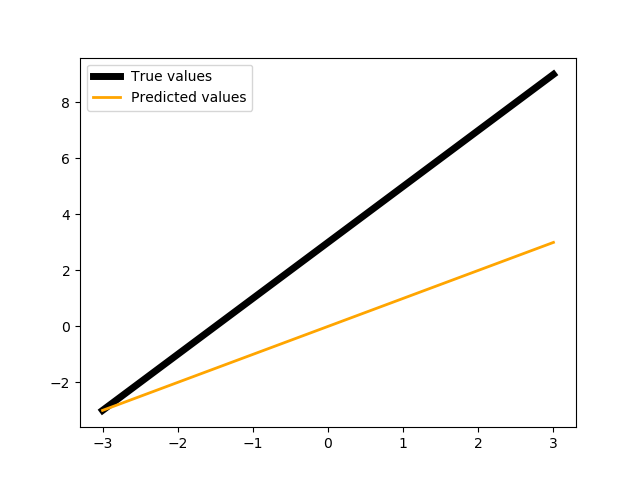
We show the values of the pseudo-neuron:
show(X, Y, neuronPL(to_gpu(X)).get())
Finally, we will train both neurons:
optimizer.learnRate = learnRate
for i in range(steps):
idx = np.random.randint(0, 1000)
x = X[idx]
y = f(x).astype(np.float32)
perceptron.optimize(x, y, learnRate)
cost.resetAccumulator()
optimizeModule(neuronPL, cost, optimizer, x, y)
neuronPL.evalMode()
showSubplots(
X,
Y,
{
"y": [perceptron(x) for x in X],
"name": "Neuron",
"color": "orange"
},
{
"y": neuronPL(to_gpu(X)).get(),
"name": "PuzzleLib neuron",
"color": "magenta"
}
)
trainBoth(500)
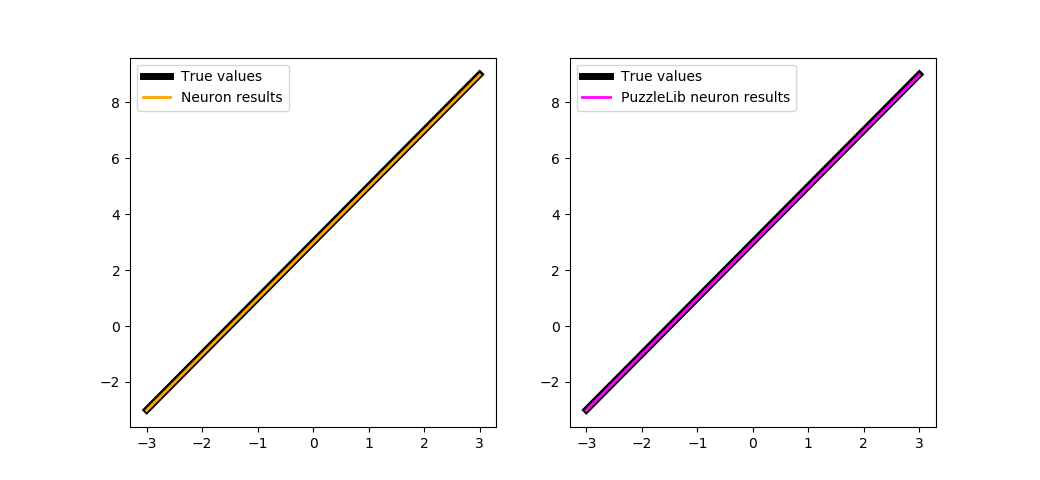
Please pay attention that the library provides handlers, which means that we do not have to create manually functions like optimizeModule, besides, they provide training on batches. If you rewrite the trainBoth function using a handler, you will get:
def trainBoth(steps=1000, learnRate=1e-2):
from PuzzleLib.Modules import Linear
from PuzzleLib.Optimizers import SGD
from PuzzleLib.Cost import MSE
from PuzzleLib.Handlers import Trainer
from PuzzleLib.Backend.gpuarray import to_gpu
neuronPL = Linear(insize=1, outsize=1)
neuronPL.W.fill(1)
neuronPL.b.fill(0)
cost = MSE()
optimizer = SGD(learnRate)
optimizer.setupOn(neuronPL)
trainer = Trainer(neuronPL, cost, optimizer, batchsize=1)
perceptron = Neuron()
show(X, Y, neuronPL(to_gpu(X)).get())
for i in range(steps):
idx = np.random.randint(0, 1000)
x = X[idx]
y = f(x).astype(np.float32)
perceptron.optimize(x, y, learnRate)
trainer.trainFromHost(x.reshape(-1, 1), y.reshape(-1, 1), macroBatchSize=1,
onMacroBatchFinish=lambda train: print("PL module error: %s" % train.cost.getMeanError()))
neuronPL.evalMode()
showSubplots(
X,
Y,
{
"y": [perceptron(x) for x in X],
"name": "Neuron",
"color": "orange"
},
{
"y": neuronPL(to_gpu(X)).get(),
"name": "PuzzleLib neuron",
"color": "magenta"
}
)
Extra: The bias role in the neuron¶
What would happen if we remove the bias parameter from the neuron? We will rewrite the neuron's methods so that there is no bias during the forward propagation:
def forward(self, data):
self.inData = data
self.data = data * self.w
return self.data
trainNeuron(500)
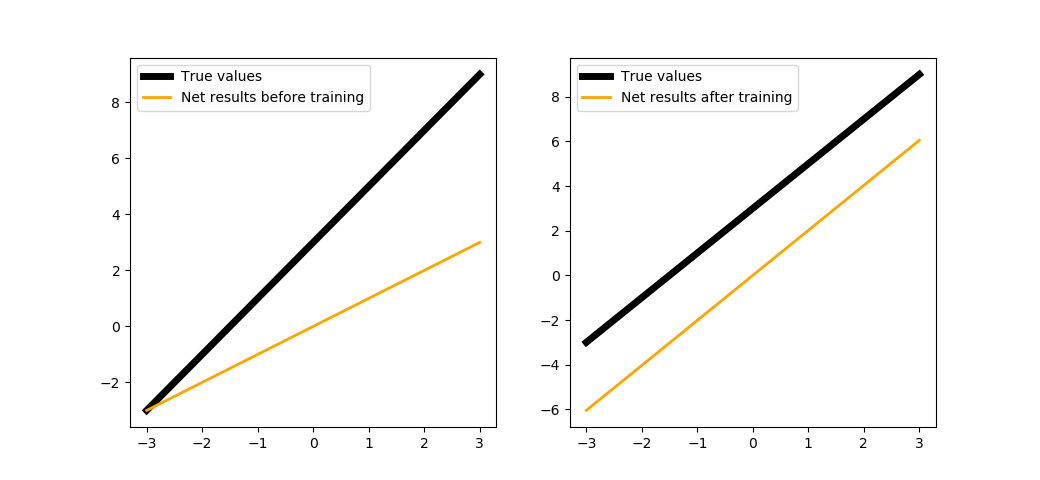
As you can see in the Fig. 8, the results provided by the neuron with no bias are in parallel with the desired function, but it lacked bias to fully restore it.